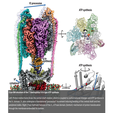
Archaeal/Vacuolar type ATP synthetase
Comparison model to pair with Bacterial ATP synthetase (https://www.thingiverse.com/thing:3729898)
Use the two models to explore A/V-type (archaeal, vacuolar) and F-type (bacterial, mitochondrial) structures.
This 3D molecular structure was derived from the original data deposited in the Protein Databank (PDB):
6QUM
• Thermus thermopiles ATP syntetase V/A type
• DOI: 10.2210/pdb6QUM/pdbEMDataResource: EMD-4640
• Classification: MEMBRANE PROTEIN
• Organism(s): Thermus thermophilus (strain HB8 / ATCC 27634 / DSM 579)
• Deposited: 2019-02-27 Released: 2019-08-28
• Deposition Author(s): Zhou, L., Sazanov, L.
Experimental Data Snapshot
• Method: ELECTRON MICROSCOPY
• Resolution: 3.25 Å
• Aggregation State: PARTICLE
• Reconstruction Method: SINGLE PARTICLE
Structure and conformational plasticity of the intact Thermus thermophilus V/A-type ATPase. Zhou, L., Sazanov, L.A.
• (2019) Science 365: --
• PubMed: 31439765
• DOI: 10.1126/science.aaw9144
• Publication Link - paywalled
• Primary Citation of Related Structures: 6R10, 6R0Z, 6R0Y, 6R0W
• Structured Abstract:
INTRODUCTION
The proton-translocating adenosine triphosphatase (ATPase)/ATP synthase family includes the F-type found in bacteria, mitochondria, and chloroplasts; the V-type (vacuolar) found in eukaryotic intracellular membrane compartments, and the A-type (archaeal) found in the plasma membrane of archaea and some eubacteria. These essential molecular machines share overall gross building plans and a rotary-catalysis mechanism. F-type and A-type enzymes function as proton motive force–driven ATP synthases, whereas V-type enzymes work as ATP-dependent proton pumps acidifying luminal environments. ATPases consist of a soluble F1/V1 domain synthesizing/hydrolyzing ATP and a membrane-embedded Fo/Vo domain translocating protons across membrane. V- and A-ATPases are similar structurally and differ from the F-ATPases by having additional peripheral stalks and connecting subunits between V1 and Vo.
Some eubacteria laterally acquired a V/A-type enzyme (including Thermus thermophilus, ThV1Vo). Rotation of the ThV1Vo membrane-embedded c12 ring relative to the stator subunit a is driven by sequential protonation and deprotonation of a conserved c subunit glutamate. Torque is transmitted via the connecting subunit d to the central stalk (DF shaft) rotating inside the V1 domain, resulting in ATP synthesis due to sequential conformational changes in subunits A3B3. The A3B3 domain itself is prevented from rotation by its connection to the two peripheral stalks.
RATIONALE
High-resolution structures of the intact ATP synthases are known for F-type enzymes, whereas there are only models of the isolated V1 and Vo domains for V-type ATPases. The details of V1-Vo coupling are thus not understood. We activated ThV1Vo by thermal cycling and reconstituted it into lipid nanodiscs for structural characterization by cryo–electron microscopy (cryo-EM).
RESULTS
We determined structures of ThV1Vo at an overall resolution of up to 3.5 Å in three main rotational states and two substates of the lowest energy state. Global conformational plasticity of ThV1Vo, as signified by asymmetric V1 precession among rotational states, is a result of mechanical competition between rotation of the bent central rotor and stiffness of the stator. One adenosine diphosphate (ADP) is observed at the ABclosed interface, with the ABsemi interface void of nucleotide, indicating that the activation process removed one inhibitory ADP molecule. An additional putative inhibitory ADP′ molecule is found between the ABclosed and D subunits. ThV1-Vo coupling is achieved via close structural match between the DF shaft and subunit d, as well as electrostatic interactions between subunit d and the c12 ring. Visualization of the proton path in Ao reveals that A-ATPases lack one of the two key glutamates found in proton access half-channels of subunit a in eukaryotic V-ATPases.
CONCLUSION
Our structure of the intact V/A-type enzyme reveals how Vo and V1 domains are coupled and serves as a reference for the entire V-type ATPase family. The additional peripheral stalk in V/A-type ATP synthase helps to control the stator subunit a position when coupling c-ring rotation to one-off 120° rotation in V1. This is in contrast to mammalian F-ATP synthase with a single peripheral stalk, where F1 rotation is divided into several smaller substeps. Threefold symmetry in the c12 ring is a feature of ThV1Vo, which necessitates nonsynchronicity between the ThV1 and Vo dwell positions to avoid local low-energy minima. Our resolved substates demonstrate the central stalk flexibilities required to accommodate such rotational mismatch. This structure fills in the gap in the ATPase evolutionary tree.
Organizational Affiliation:
Institute of Science and Technology Austria, Klosterneuberg 3400, Austria. sazanov@ist.ac.at.,Institute of Science and Technology Austria, Klosterneuberg 3400, Austria.

/https://fbi.cults3d.com/uploaders/29629622/illustration-file/975fab55-4fb2-44a8-ba91-fae904e02d06/20191012_101607.jpg)